- Search
Abstract
Purpose
Oxaliplatin is a third-generation platinum compound, and it has no nephrotoxicity and has reduced bone marrow toxicity. Cancer cells that are resistant to cisplatin are sensitive to oxaliplatin. Oxaliplatin is used widely for the treatment of colon cancers. Recently, oxaliplatin was reported to inhibit the expression of survivin, which protects cell apoptosis. However, there are no reports on the expressions of survivin variants and the changes in intracellular localization of survivin in cancer cells. We studied the expression of survivin caused by oxaliplatin in HCT116 colon cancer cells, and we observed the localization of survivin in the mitotic phase.
Methods
We treated the HCT116 colon cancer cells with 2.0 µM of oxaliplatin, and we studied the expressions of survivin protein, and survivin mRNA variants, as well as the changes in intracellular localization, by using the Western blot method, RT-PCR, immunocytochemistry, and flowcytometry.
Results
Oxaliplatin inhibits the expression of the survivin protein and survivin mRNA in HCT116 colon cancer cells. The expression of the survivin-2B variants, which have no antiapoptotic activity but control cell mitosis by localization on a microtubule, is reduced continuously 2 days after treatment with oxaliplatin. In immunocytochemistry, expression of survivin in the cytoplasm is reduced and especially is not expressed in microtubules and contractile rings.
The incidence of colon cancer ranks 4th among males and 3rd among females in Korea. Ninety percent of the patients with colon cancer are cured through surgical excision, but approximately half of the patients experience relapse or metastasis to other organs after the treatment [1]. Relapsing colon cancer is treated with anticancer agent primarily, but the patient gains tolerance to specific drugs during treatment.
Survivin is a type of inhibitor of apoptosis (IAP) protein that has the size of 16 kD and helps repress apoptosis. There are three different types of survivin with alternative splicing variants: wild-type survivin, survivin-2B (with duplicated 2nd axon), and survivin-ΔEx3 (with missing 3rd axon). These variations are known to have different functions. Generally, survivin-ΔEx3 contains the apoptosis repressing function, but survivin-2B is known to have lost the apoptosis repressing function [2]. In addition, it was revealed that the 3 different genes of survivin exist in different intracellular location of different cells. However, the functions of each survivin in cancer are not yet discovered [3].
Because cisplatin and carboplatin, platinum compound drugs, are less effective to colon cancer, they are administrated either singularly or in combination with 5-fluorouracil (5-FU) and shows approximately 20% response [4]. Oxaliplatin is a drug based on 3rd generation platinum compound which was developed after the discovery of cisplatin and carboplatin. Oxaliplatin was reported to have antitumor effect on cell strains that are resistant to cisplatin and carboplatin [5]. Acting mechanism why oxaliplatin shows stronger cytotoxicity to cancer cell that has resistance to the other platinum compound drugs and why it is more effective when combined with other anticancer drugs are not known [6]. However, repressing expression of survivin that suppress the apoptosis was reported to be one of the mechanisms of the oxaliplatin [7].
The authors of this research focused on oxaliplatin's effect on the change of expression of survivin variants and whether oxaliplatin shows cytotoxicity by measuring change of expression of survivin protein and each variants of survivin mRNA.
Oxaliplatin was donated by Sanofi-Synthelabo Korea (Seoul, Korea), and the optimum concentration of oxaliplatin was determined using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) method. One-tenth, 0.2, 0.5, 1.0, 2.0, 5.0, and 10 µM of oxaliplatin were each added to separate HCT116 colon cancer cell strain samples and were cultured for 48 hours. Fresh culture fluid containing corresponding amounts of oxaliplatin was replaced each day, and after 48 hours after culture MTS reagent was added to the cells. The mixtures were settled for 2 hours. Cytotoxicity data were collected by measuring the optical density with an enzyme linked immunosorbent assay (ELISA) reader.
Oxaliplatin was diluted to 2.0 µM and was added to the HCT116 colon cancer cell strain, and the samples were cultured for 1, 2, and 3 days. Fresh culture media containing corresponding amounts oxaliplatin were used each day. After 48 hours, cells were dissolved in cell lysis buffer solution to extract the protein. Fifty µg of protein was separated through electrophoresis in 3% polyacrylamide gel. This protein was transferred into polyvinylidene fluoride (PVDF), a Western blot was executed with a specific survivin antibody (anti-hSurvivin, R&D Systems, polyclonal, Cat. NO. AF886, Minneapolis, MN, USA), and reacted bands were detected with an image analyzer (LAS-2000, Fuji Film, Tokyo, Japan).
Two µM of oxaliplatin was added to the HCT116 colon cancer cell strain and was cultured for 1, 2, and 3 days. Then, Trizol reagent (Life Technologies, Rockville, MD, USA) was used to extract total RNA. Cells that were cultured were dissolved in 1 mL of cold Trizol solution and lysed. Then, they went through the centrifuge at 12,000 g at 4℃ for 10 minutes, and the supernatant was moved into a new eppendorf tube and was then settled at room temperature for 5 minutes. Chloroform, 200 µL, was added, thoroughly agitated for 15 seconds, and centrifuged at 12,000 g at 4℃ for 15 minutes after being settled for 3 minutes at room temperature. Supernatant-containing RNA was carefully moved to a new eppendorf tube, and 500 µL of isopropyl alcohol was added and centrifuged at 12,000 g at 4℃ for 10 minutes after being settled at room temperature for 10 minutes. Supernatant was removed after checking the presence of RNA clusters, and 1 mL of 75% ethanol was added to clean it. Then, it was centrifuged at 7,500 g at 4℃ for 5 minutes, ethanol was removed, and the RNA was obtained after drying it in the air for 15 minutes. This RNA was dissolved in secondary distilled water that was treated with diethyl pyrocarbonate (DEPC, Sigma Chemical Co. St Louis, MO, USA), and the concentration of RNA was measured with a spectrophotometer measuring the optical density at 260/ 280 nm. The separated RNA was stored at -80℃.
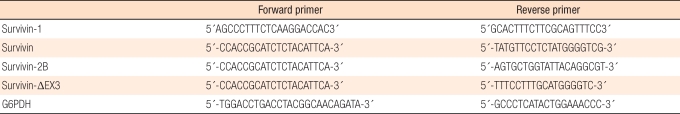
In creating the cDNA, 2 µL of Oligo dT (Promega, Madison, WI, USA) and secondary distilled water that was treated with DEPC were added to 5 µg/µL of the RNA obtained from the HCT116 colon cancer cell strain until the mixture was 10 µL. Then, a fixation reaction was allowed to occur for 10 minutes at 70℃, the mixture was then cooled with ice for 10 minutes, and a master mix (5 × first strand buffer, 4 µL; 0.1 M DTT, 2 µL; 2.5 M dNTP, 2 µL; RNase inhibitor, 1 µL) was added. The mixture was reacted for 2 minutes at 42℃, 1 µL of reverse transcriptase (superscript II [Life Technologies] was added, and the mixture was reacted again for 50 minutes at 42℃ and 15 minutes at 70℃ to finally obtain the cDNA. Secondary distilled water without nuclease, 36.5 µL, and 5 µL of master mix (10 × buffer [500 mM KCl, 100 mM Tris-HCl [pH 8.3], 15 mM MgCl2]), 4 µL of dNTP, 1 µL of primer sense, 1 µL of primer antisense, and 0.5 µL of Taq DNA polymerase (TaKaRa Taq™, Takara, Shiga, Japan) were added to 2 µL of cDNA. Then, the mixture was placed in a DNA thermal cycler (GeneAmp®PCR system 9700, Applied Biosystems, Foster City, CA, UAS) for 2 minutes at 94℃, 1 minutes (denaturing) at 94℃, 1 minutes (annealing) at 60℃, and 1 minutes (extension) at 72℃. Under the conditions of PCR, the process was repeated 28 times to amplify the reaction. After the PCR reaction, electrophoresis was executed in 1.5% agarose gel, and GAPDH was modified by using the same method. The primer for survivin-splicing variants is shown in Table 1 [8].
In order to find the optimum concentration of oxaliplatin, the cytotoxicity of oxaliplatin for the HCT116 colon cancer cell strain was measured using the MTS method. The IC50 concentration was 2.0 µM in oxaliplatin, which showed the cytotoxicity to be approximately 50% of the cells (Fig. 1). Forty-eight hours after each concentration of oxaliplatin had been added to the HCT116 colon cancer cell strain, the cultured cells were observed through a phase-contrast microscope (Fig. 2). When compared with the control group (no oxaliplatin added), the culture with a 1.0 µM concentration of oxaliplatin began to show a reduced cell count; cultures with 5.0 µM or higher concentrations showed noticeable reductions in cell count, and the cells were also deformed. Therefore, a 2.0 µM dose oxaliplatin was used in all the experiment because it was considered to be the most appropriate dose.
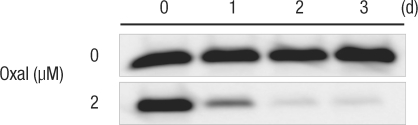
Two µM of oxaliplatin was added to the HCT116 colon cancer cell strain and the culturing results were collected after three days. Survivin protein expression was reduced 1 day after the oxaliplatin was added, and no survivin protein was expressed after the second day (Fig. 3).
The RT-PCR method was used to measure the expression of the total survivin mRNA and of each variant of mRNA. For the onset of survivin mRNA in variant forms caused by the primers, each specific primer was used and showed a single RT-PCR band. This showed that each primer was appropriate in testing the onset of variant mRNA (Fig. 4). When compared with the control group without oxaliplatin, survivin wild-type mRNA showed reduced expression on the second day, but the expression increased again on day 3. Survivin-2B variant did not show any change on the first day, but showed reduced expression throughout days two and three. Survivin-ΔEx3 variant had reduced expression from the first day, in spite of its small amount (Fig. 5).
After treatment with oxaliplatin for 24 hours and 48 hours, the cell-cycle changes in both the control group and the treated group were measured using flowcytometry. After 24 hours of culture, the percent of subG1 phase (representing the death of a cell) cells increased from 12.47% to 22.39%. After 48 hours, it increased from 9.47% to 28.21%. The percent of cells in the G0-G1 (gap 0 and gap1) phase decreased from 82.56% to 67.65% after 24 hours and from 80.50% to 59.76% after 48 hours. The S (DNA synthesis) phase fraction increased from 5.10% to 5.66% after 24 hours, reduced from 9.32% to 5.25% after 48 hours, and showed a slight increase after that. The percent of cells in the G2 (gap 2) phase increased from 0.25% to 4.61% after 24 hours and from 0.36% to 5.04% after 48 hours (Fig. 6).
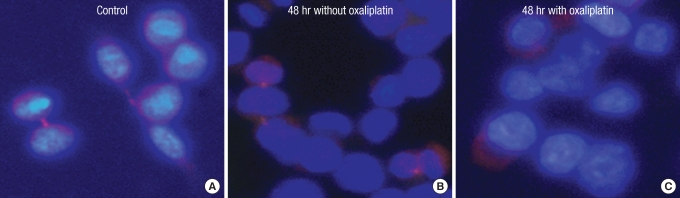
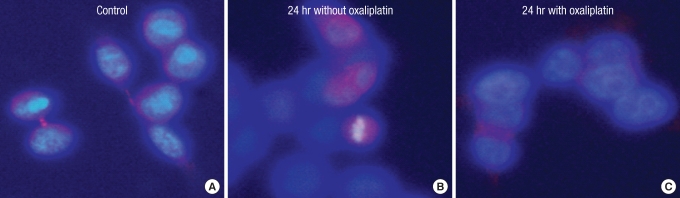
When the sample is cultured without oxaliplatin, the survivin protein often appears to be located in the cytoplasm because the contractile ring or microtubule areas are lightly dyed (Figs. 7A-C, 8A, 9A). After culturing the control group for 24 hours, the protein is thought to be located in the chromosomes or the microtubule because it gets dyed with the chromosomes during the mid-phase of mitosis (Figs. 7D-F, 8B). It gets dyed lightly in the cytoplasm after 48 hours of culture and is thought to be located in the contractile ring or microtubule during cytokinesis (Figs. 7G-I, 9B). However, after treatment with oxaliplatin for 24 hours and 48 hours, only the cytoplasm gets lightly dyed, and the survivin protein's intracellular change of location is thought to be caused by oxaliplatin (Figs. 7J-O, 8C, 9C).
Survivin is one of the human IAP homologues, and it belongs to the inhibitor of apoptosis protein (IAP) family [9]. Every single IAP has one or more baculovirus IAP repeat (BIR), and a BIR consists of approximately 70 amino acids [10]. Eight human IAPs, c-IAP1, c-IAP2, XIAP, NAIP, survivin, apollon, ML-IAP/livin, ILP-2, have been found [11]. Survivin is located at 17q25, consists of 142 amino acids, and has the size of 16.5 kDa. Survivin has the structure of a long alpha-helix, does not have the RING finger, and has a single BIR area structurally. The long alpha-helix structure of the carboxyl terminal in survivin interacts with the microtubules [12]. A gene of the survivin has 4 main axons (1, 2, 3, and 4) and 2 hidden axons (2B and 3B). Accordingly, there are three different variants of survivin, survivin-2B, survivin-ΔEx3, and survivin-3B. Survivin-ΔEx3 is a variant with the loss of the third axon, and survivin-2B is a variant with an additional short axon called intron 2.
The expression of survivin is controlled by the cell cycle and is related to the control of cell death and cell division [13]. According to recent studies, survivin is also found in a normal colon mucosa and a proliferative polyp [14]. According to the existing research, survivin expression among patients with cancer is reported to be a bad clinical sign with an unfavorable prognosis for progression of the illness and a reduced survival rate. Thus, survivin detection in urine for patients with bladder cancer is considered to be a sign for an unfavorable prognosis, and a urine test for survivin detection is accredited as a prognosis-predicting factor bn[15, 16]. In stomach cancer, the expression of survivin was reported to indicate an unfavorable prognosis and reduced cell death [17]. However, other studies reported that although expression of survivin within the nucleus is related to a favorable prognosis, expression within the cytoplasm is not related with an unfavorable prognosis [18]. It was also reported that positive nucleus dyeing in hepatic cancer is related to proliferation [19]. Expression of survivin in colorectal cancer is related to the onset of bcl-2, tumor growth, and low survival rate. It is also related to the response in chem otherapy of patients with rectal cancer [20-25]. Among survivin variants, survivin-ΔEx3 is known to have relationships with apoptosis, but the reduction of survivin-2B onset is known to have relationships with the progress of cancer in stomach cancer, colon cancer, and many other types of cancer [8, 26-29].
Oxaliplatin is a primary anticancer drug that is used in patients with stage 3 or higher colon cancer or metastatic colorectal cancer. It is in the family of 3rd-generation platinum compound anticancer drugs, but does not have nephrotoxicity related to cisplatin, has less myelosuppression related to carboplatin, and has a higher response rate to colon cancer when compared to cisplatin. However, there is not much known about the precise acting mechanism of oxaliplatin, and recent studies have reported the cytotoxicity by repressing survivin expression to be one of the acting mechanism [5-7].
This research observed that survivin protein expression was reduced one day after the oxaliplatin was added to the colon cancer cell strain. The expression started to decrease after the first day and was almost completely repressed after the second day. Also, according to the RT-PCR measurements of the survivin variant expression rate with its special primer, wild-type survivin mRNA showed decreased expression after the second day, but showed a recovery after the third day. Among survivin variants, the expression of survivin-2B (known for not repressing cell death) was reduced after the second day and kept on decreasing after the third day. The survivin-ΔEx3 variant (with missing 3rd axon) showed reduced expression after the first day and continued to show a decrease even after the third day. The total survivin mRNA expression was decreased by oxaliplatin, but recovered again. On the other hand, expression of the survivin variant not related to apoptosis decreased continuously.
Even in the cell-cycle analysis done by using flowcytometry, cells treated with oxaliplatin and cultured for 24 hours and 48 hours had increased cell death of approximately 10% to 22-28% compared to the control sample. This shows that the expression of survivin recovers on the third day, but survivin-ΔEx3, which represses apoptosis, decreases continuously, and apoptosis occurs continuously. The reason for reduced expression of survivin-2B (with almost no repression of apoptosis) is thought to be the dislocation of the survivin-2B in the G2/M phase, which causes a mitotic catastrophe by disturbing bipolar spindle fiber formation during mitosis.
The results of dying DAPI and survivin after treating with oxaliplatin showed reduced survivin expression within the cytoplasm when compared with the control group. Also, survivin does not appear in microtubules or chromosomes during mitosis, which is thought to cause disruption in cell division. More research is necessary in order to determine which variant survivin is found within either the nucleus or the cytoplasm and which causes not only cell death but also mitotic catastrophe.
According to our results, mechanisms of oxaliplatin include repression of the onset of survivin, cell death, and lack of formation in microtubules or contractile rings during the cell cycle. Therefore, as reported by Fortugno [3], it is thought to disturb the formation of bipolar spindle fibers during mitosis.
Oxaliplatin's acting mechanism not only represses the onset of survivin in the HCT116 colon cancer cell strain but also causes apoptosis by changing the location of expression within the cell. Also, it fails to settle in the microtubules, causing disorder in mitotic recombination that will show cytotoxicity.
References
1. Nelson H, Petrelli N, Carlin A, Couture J, Fleshman J, Guillem J, et al. Guidelines 2000 for colon and rectal cancer surgery. J Natl Cancer Inst 2001;93:583–596. PMID: 11309435.


2. Mahotka C, Liebmann J, Wenzel M, Suschek CV, Schmitt M, Gabbert HE, et al. Differential subcellular localization of functionally divergent survivin splice variants. Cell Death Differ 2002;9:1334–1342. PMID: 12478470.


3. Fortugno P, Wall NR, Giodini A, O'Connor DS, Plescia J, Padgett KM, et al. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci 2002;115:575–585. PMID: 11861764.


4. Kemeny N, Israel K, Niedzwiecki D, Chapman D, Botet J, Minsky B, et al. Randomized study of continuous infusion fluorouracil versus fluorouracil plus cisplatin in patients with metastatic colorectal cancer. J Clin Oncol 1990;8:313–318. PMID: 2405107.


5. Rixe O, Ortuzar W, Alvarez M, Parker R, Reed E, Paull K, et al. Oxaliplatin, tetraplatin, cisplatin, and carboplatin: spectrum of activity in drug-resistant cell lines and in the cell lines of the National Cancer Institute's Anticancer Drug Screen panel. Biochem Pharmacol 1996;52:1855–1865. PMID: 8951344.


6. Raymond E, Faivre S, Chaney S, Woynarowski J, Cvitkovic E. Cellular and molecular pharmacology of oxaliplatin. Mol Cancer Ther 2002;1:227–235. PMID: 12467217.

7. Fujie Y, Yamamoto H, Ngan CY, Takagi A, Hayashi T, Suzuki R, et al. Oxaliplatin, a potent inhibitor of survivin, enhances paclitaxel-induced apoptosis and mitotic catastrophe in colon cancer cells. Jpn J Clin Oncol 2005;35:453–463. PMID: 16024531.


8. Meng H, Lu C, Mabuchi H, Tanigawa N. Prognostic significance and different properties of survivin splicing variants in gastric cancer. Cancer Lett 2004;216:147–155. PMID: 15533590.


9. Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 1997;3:917–921. PMID: 9256286.


10. Duckett CS, Nava VE, Gedrich RW, Clem RJ, Van Dongen JL, Gilfillan MC, et al. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J 1996;15:2685–2694. PMID: 8654366.



11. Chen Z, Naito M, Hori S, Mashima T, Yamori T, Tsuruo T. A human IAP-family gene, apollon, expressed in human brain cancer cells. Biochem Biophys Res Commun 1999;264:847–854. PMID: 10544019.


12. Chantalat L, Skoufias DA, Kleman JP, Jung B, Dideberg O, Margolis RL. Crystal structure of human survivin reveals a bow tieshaped dimer with two unusual alpha-helical extensions. Mol Cell 2000;6:183–189. PMID: 10949039.


14. Gianani R, Jarboe E, Orlicky D, Frost M, Bobak J, Lehner R, et al. Expression of survivin in normal, hyperplastic, and neoplastic colonic mucosa. Hum Pathol 2001;32:119–125. PMID: 11172305.


15. Swana HS, Grossman D, Anthony JN, Weiss RM, Altieri DC. Tumor content of the antiapoptosis molecule survivin and recurrence of bladder cancer. N Engl J Med 1999;341:452–453. PMID: 10438269.

16. Smith SD, Wheeler MA, Plescia J, Colberg JW, Weiss RM, Altieri DC. Urine detection of survivin and diagnosis of bladder cancer. JAMA 2001;285:324–328. PMID: 11176843.


17. Wakana Y, Kasuya K, Katayanagi S, Tsuchida A, Aoki T, Koyanagi Y, et al. Effect of survivin on cell proliferation and apoptosis in gastric cancer. Oncol Rep 2002;9:1213–1218. PMID: 12375022.


18. Okada E, Murai Y, Matsui K, Isizawa S, Cheng C, Masuda M, et al. Survivin expression in tumor cell nuclei is predictive of a favorable prognosis in gastric cancer patients. Cancer Lett 2001;163:109–116. PMID: 11163114.


19. Ito T, Shiraki K, Sugimoto K, Yamanaka T, Fujikawa K, Ito M, et al. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology 2000;31:1080–1085. PMID: 10796883.


20. Kawasaki H, Altieri DC, Lu CD, Toyoda M, Tenjo T, Tanigawa N. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res 1998;58:5071–5074. PMID: 9823313.

21. Yamamoto T, Manome Y, Nakamura M, Tanigawa N. Down regulation of survivin expression by induction of the effector cell protease receptor-1 reduces tumor growth potential and results in an increased sensitivity to anticancer agents in human colon cancer. Eur J Cancer 2002;38:2316–2324. PMID: 12441269.


22. Chen WC, Liu Q, Fu JX, Kang SY. Expression of survivin and its significance in colorectal cancer. World J Gastroenterol 2004;10:2886–2889. PMID: 15334693.



23. Knutsen A, Adell G, Sun XF. Survivin expression is an independent prognostic factor in rectal cancer patients with and without preoperative radiotherapy. Int J Radiat Oncol Biol Phys 2004;60:149–155. PMID: 15337550.


24. Capalbo G, Rödel C, Stauber RH, Knauer SK, Bache M, Kappler M, et al. The role of survivin for radiation therapy. Prognostic and predictive factor and therapeutic target. Strahlenther Onkol 2007;183:593–599. PMID: 17960333.


25. Baek MJ, Lee EM, Kim CJ, Park NK, Shin EJ, Jang YS, et al. Expression of Survivin and Its Correlation with Prognosis in Colorectal Cancer. J Korean Soc Coloproctol 2006;22:125–131.
26. Islam A, Kageyama H, Hashizume K, Kaneko Y, Nakagawara A. Role of survivin, whose gene is mapped to 17q25, in human neuroblastoma and identification of a novel dominant-negative isoform, survivin-beta/2B. Med Pediatr Oncol 2000;35:550–553. PMID: 11107115.


27. O'Driscoll L, Linehan R, M Kennedy S, Cronin D, Purcell R, Glynn S, et al. Lack of prognostic significance of survivin, survivin-deltaEx3, survivin-2B, galectin-3, bag-1, bax-alpha and MRP-1 mRNAs in breast cancer. Cancer Lett 2003;201:225–236. PMID: 14607338.


28. Suga K, Yamamoto T, Yamada Y, Miyatake SI, Nakagawa T, Tanigawa N. Correlation between transcriptional expression of survivin isoforma and clinicopathological findings in human colorectal carcinomas. Oncol Rep 2005;13:891–897. PMID: 15809755.

29. He XF, Wen DG, Hou JQ, He J, Cen JN. Expression of survivin and the splice variants survivin-2B and survivin-ΔEx3 in bladder cancer and their clinical significance. Ai Zheng 2009;28:1209–1213. PMID: 19895744.


Fig. 1
Cytotoxi city of oxaliplatin for HCT116 colon cancer cells, as determined by using the 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)- 2-(4-sulfophenyl)-2H-tetrazolium (MTS) method.
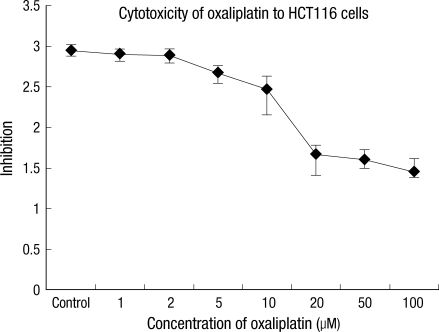
Fig. 3
Phase contrast images of HCT116 colon cancer cells after incubation with various concentration of oxaliplatin for 48 hours.

Fig. 4
Survivin mRNA expression in HCT116 colon cancer cells when using survivin variant-specific primers.
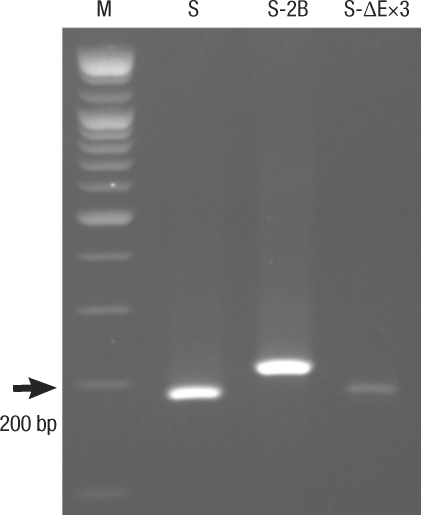
Fig. 5
Survivin variant mRNA expression in HCT116 colon cancer cells after incubation with 2.0 µM of oxaliplatin for 1-3 days.
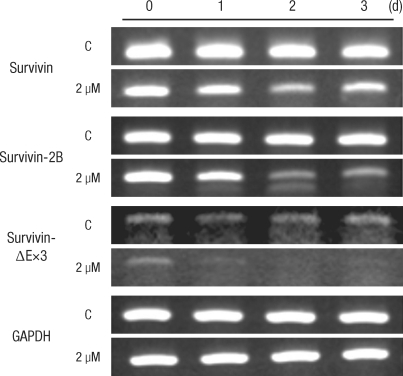
Fig. 6
Measurement of the cell-cycle phase by using flowcytometry after incubation with 2.0 µM of oxaliplatin for 24 and 48 hours.
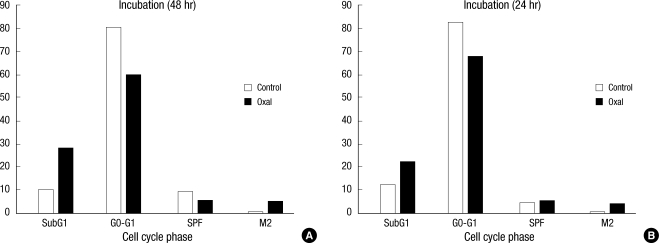
Fig. 7
Intracellular localization change of survivin after incubation with 2.0 µM of oxaliplatin. HCT116 colon cancer cells are stained with DAPI (A, D, G, J, M), survivin (B, E, H, K, N), and merge (C, F, I, L, O). We cultured the HCT116 cells for a control (A-C), without oxaliplatin for 24 hours (D-F) and 48 hours (G-I), and with 2.0 µM of oxaliplatin for 24 hours (J-L) and 48 hours (M-O).
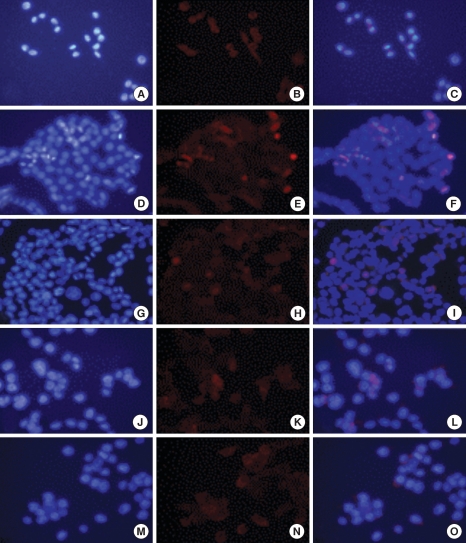
Fig. 8
Intracellular localization change of survivin after incubation for 24 hours without or w 2.0 µM of oxaliplatin.
- TOOLS