Performance reporting design in artificial intelligence studies using image-based TNM staging and prognostic parameters in rectal cancer: a systematic review
Article information
Abstract
Purpose
The integration of artificial intelligence (AI) and magnetic resonance imaging in rectal cancer has the potential to enhance diagnostic accuracy by identifying subtle patterns and aiding tumor delineation and lymph node assessment. According to our systematic review focusing on convolutional neural networks, AI-driven tumor staging and the prediction of treatment response facilitate tailored treatment strategies for patients with rectal cancer.
Methods
This paper summarizes the current landscape of AI in the imaging field of rectal cancer, emphasizing the performance reporting design based on the quality of the dataset, model performance, and external validation.
Results
AI-driven tumor segmentation has demonstrated promising results using various convolutional neural network models. AI-based predictions of staging and treatment response have exhibited potential as auxiliary tools for personalized treatment strategies. Some studies have indicated superior performance than conventional models in predicting microsatellite instability and KRAS status, offering noninvasive and cost-effective alternatives for identifying genetic mutations.
Conclusion
Image-based AI studies for rectal cancer have shown acceptable diagnostic performance but face several challenges, including limited dataset sizes with standardized data, the need for multicenter studies, and the absence of oncologic relevance and external validation for clinical implantation. Overcoming these pitfalls and hurdles is essential for the feasible integration of AI models in clinical settings for rectal cancer, warranting further research.
INTRODUCTION
Over 1 million people worldwide die due to colorectal cancer (CRC) each year. According to the statistics by the US National Cancer Institute, CRC is the third most common cancer in both men and women [1]. Despite advancements in treatment and implementation of a nationwide screening program, the mortality rate has steadily increased [2]. The current status of treatment of patients with rectal cancer requires a multidisciplinary strategy encompassing local excision, total mesorectal excision (TME), chemotherapy, and radiotherapy [3]. For patients with early rectal cancer, local excision is an optional treatment within the spectrum of organ-preserving strategies [4, 5]. In patients with advanced rectal cancer, TME is the treatment of choice [6, 7]. Recently, minimally invasive surgical techniques, such as robotic surgery and transanal surgery, have been increasingly performed [8, 9]. In patients with advanced rectal cancer, if there is a good response to treatment after preoperative chemoradiotherapy (CRT), local excision or a nonoperative management, also known as the “watch-and-wait” strategy, may be attempted [10, 11]. It is important to select the appropriate candidate for tailored treatment in rectal cancer, which requires accurate diagnosis and assessment of response to preoperative treatment [12, 13].
Magnetic resonance imaging (MRI) is considered to be the most valuable imaging modality for primary staging and restaging after CRT, guiding subsequent medical decisions in rectal cancer management [14–18]. With advancements in artificial intelligence (AI), researchers and clinicians have been exploring innovative ways to utilize MRI data through AI algorithms to improve diagnostic accuracy and prognostic prediction. By harnessing the power of AI, it is possible to analyze MRI images with high speed and accuracy [19]. AI algorithms can identify subtle patterns within images, aiding in the precise delineation of tumor borders, assessment of lymph node involvement, and evaluation of potential metastases. Accurate staging through AI-driven imaging diagnosis can enable tailored treatment strategies, optimizing the selection of patients for neoadjuvant therapies and guiding clinicians in making decisions regarding the extent of surgical resection [20, 21]. Radiomics, an AI method, aims to establish models that enhance diagnostic accuracy by extracting and analyzing first- and high-order features from medical images [22]. Radiomics has advantages for small datasets; however, it may not perform as effectively as neural networks, particularly for large datasets [22, 23]. One of the most commonly used neural network architectures in medical imaging is a convolutional neural network (CNN). CNNs consist of convolutional, pooled, and fully connected layers. CNNs progressively identify abstract and intricate features, making them pivotal in medical imaging research for tasks such as the differential diagnosis of tumors, tumor segmentation, lesion detection, and accelerated imaging [23, 24].
With the rapid advancement of AI in medical imaging of rectal cancer, this study aimed to provide a summary of several important topics in current image-based studies using AI for patients with rectal cancer by investigating the performance reporting design based on the quality of the dataset, annotation with ground-truth labelling, model performance, and external validation. In this study, we focused on how neural networks see images identical to images clinicians see during examination. We excluded studies using radiomics to transform actual images.
METHODS
This study analyzes the utilization of images, architectures employed, dataset constitution, annotation, and diagnostic performance of each research investigation. We reviewed tumor segmentation, TNM staging, genotyping, risk factors such as circumferential resection margin (CRM), and treatment response corresponding to the current guidelines for rectal cancer treatment [15, 25]. A systematic search of the Cochrane Library, PubMed (MEDLINE), Embase, and IEEE Xplore databases was performed for studies published between 2017 and 2023. This study focused on neural network models for medical images, and excluded studies on surgical procedures using only radiomics.
RESULTS
AI in rectal cancer imaging
Segmentation
As AI technologies continue to advance, researchers have begun to explore the feasibility of applying AI techniques to tumor segmentation. Because of individual variations in the perception of the disease, manual labelling may be subjective and time-consuming. AI-based tumor segmentation can be more objective than manual labelling and can reduce labor burden. Although MRI provides clear delineation of rectal structures and tumor appearance, accurate segmentation is challenging because of the complex background of rectal images [23].
Table 1 summarizes AI studies on segmentation of rectal cancer [26–35]. A CNN was initially applied to the segmentation of rectal cancer. Trebeschi et al. [26] used a CNN model based on T2-weighted image (T2WI) and diffusion-weighted image (DWI) for 132 patients with rectal cancer. The performance of the AI model achieved a dice similarity coefficient (DSC) of 0.70 and an area under the receiver operating characteristic curve (AUROC) of 0.99. As an excellent segmentation framework for medical images, U-Net [36] has been applied for the segmentation of rectal cancer. Wang et al. [27] used a 2-dimensional (2D) U-Net based on T2WI obtained from 113 patients with rectal cancer. The DSC, Hausdorff distance, average surface distance, and Jaccard index values were 0.74, 20.44, 3.25, and 0.60, respectively. Kim et al. [28] obtained MRI of 133 patients with rectal cancer. The ground truth of the tumor for all 2D MRI was defined by 2 gastrointestinal radiologists, and 2D slices were manually selected by gastrointestinal radiologists. U-Net [36], FCN-8 (fully convolutional networks, 8 pixels) [37], and SegNet [38] were used for tumor segmentation, and their performances were compared. U-Net was superior to the other models and achieved a DSC of 0.81, sensitivity of 0.79, and specificity of 0.98. Pang et al. [29] used U-Net based on T2WI obtained from 134 patients with rectal cancer and externally validated 34 patients from different hospitals. The DSC, sensitivity, and specificity values were 0.95, 0.97, and 0.96, respectively. Knuth et al. [30] collected 2 cohorts of patients with rectal cancer from different hospitals and used 2D U-Net to perform tumor segmentation on T2WI. The DSC was 0.78. DeSilvio et al. [31] used region-specific U-Net and multiclass U-Net based on T2WI obtained from 92 patients with rectal cancer and performed external validation with 11 patients from different hospitals. The performance of the region-specific U-Net was superior to that of the multiclass U-Net. The region-specific U-Net achieved a DSC of 0.91 and a Hausdorff distance of 2.45. Researchers have used models other than U-Net. Zhang et al. [32] used a 3D V-Net [39] based on T2WI and DWI obtained from 202 patients with rectal cancer. The DSC of T2WI was 0.89±0.21 and that of DWI was 0.96±0.06. Fig. 1 shows examples of tumor segmentation of AI model [32].

Examples of rectal cancer segmentation. Illustration of automated segmentation using 3-dimensional V-Net versus ground truth on rectal magnetic resonance images of a 51-year old man. Purple indicates tumor, yellow indicates normal rectal wall, and blue indicates lumen. Reprinted from Zhang et al. [32], available under the Creative Commons Attribution License.
Studies have also been conducted on lymph node segmentation using AI. Zhao et al. [33] developed an AI model for lymph node detection and segmentation based on multiparametric MRI scans obtained from 293 patients with rectal cancer. For lymph node detection, the AI model achieved a sensitivity, positive predictive value (PPV), and false-positive rate per case of 0.80, 0.74, and 8.6 in internal testing, and 0.63, 0.65, and 8.2 in external testing. The detection performance was superior to that of junior radiologists with less of 10 years of experience. For lymph node segmentation, the DSC was in the range of 0.81 to 0.82. Fig. 2 shows examples of lymph node segmentation of AI model [33].
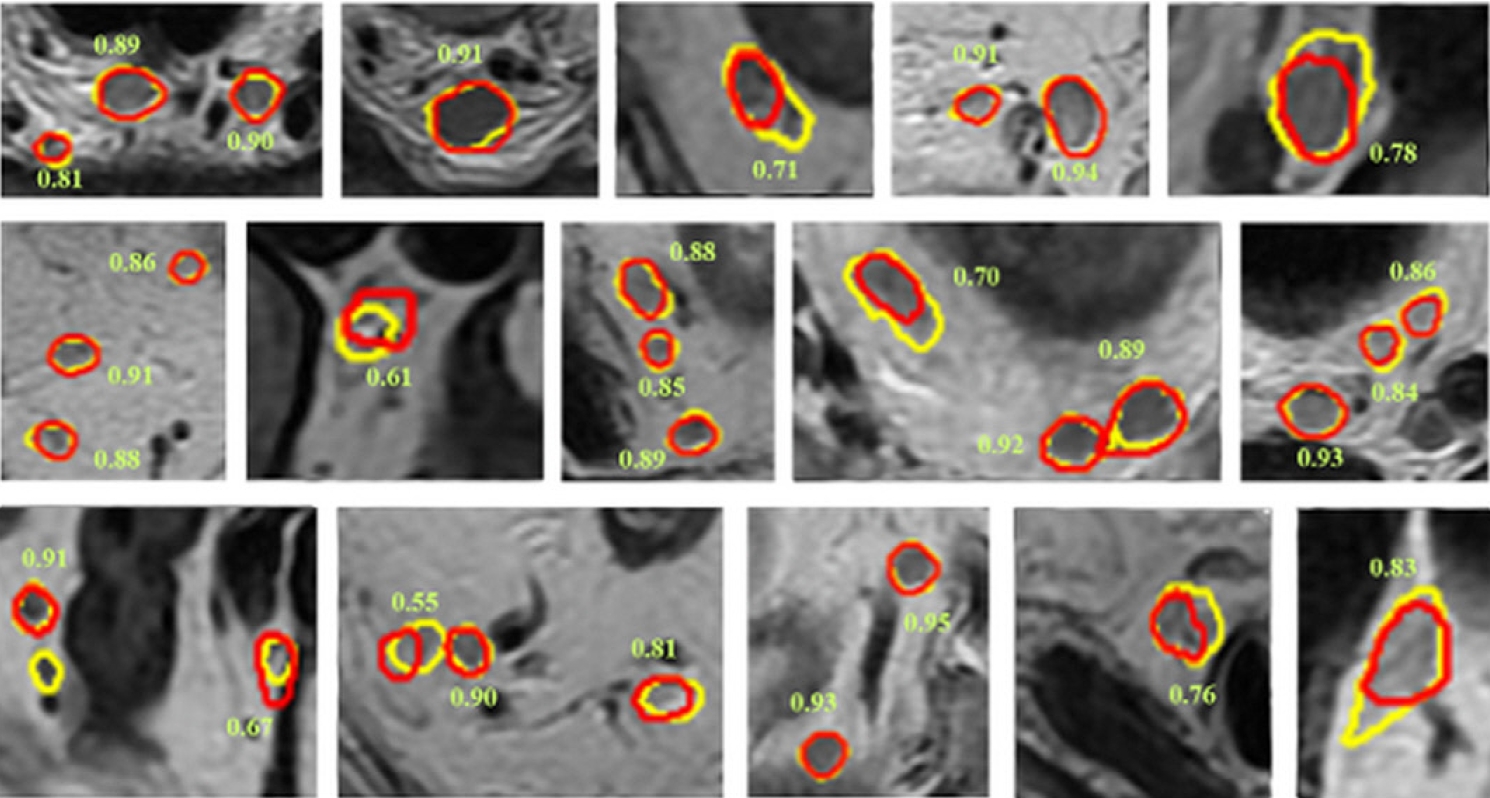
Examples of lymph node segmentation. Ground truth results are shown in yellow, and segmentation results by the artificial intelligence model are shown in red. The number besides the lymph node is the corresponding dice similarity coefficient. Reprinted from Zhao et al. [33], available under the Creative Commons License.
Because manual annotation is a time-consuming and labor-intensive task, interest in AI segmentation models has increased because artificial intelligence segmentation models have achieved good performance. Through continuous iteration and optimization of the automatic segmentation model, radiologists can be effectively assisted for faster and more accurate annotation [23]. Jian et al. [34] used U-Net and VGG-16 [40] based on T2WI obtained from 512 patients with colorectal cancer. VGG-16 was used as the base model to extract features from tumor images, and 5 side-output blocks were used to obtain accurate tumor segmentation results. After cropping the region of interest, the VGG-16 model performed automatic segmentation without intervention. The segmentation performance of VGG-16 was superior to that of U-Net. The VGG-16 achieved a DSC, PPV, specificity, sensitivity, Hammoude distance, and Hausdorff distance of 0.84, 0.83, 0.97, 0.88, 0.27, and 8.2, respectively. Zhu et al. [35] used a 3D U-Net based on DWI obtained from 300 patients with rectal cancer. The region of the rectal tumor was delineated using the DWI by experienced radiologists as the ground truth. Automatic segmentation was compared with semiautomatic segmentation. Semiautomatic segmentation first requires manually assigning a threshold value for voxel selection and then automatically segmenting the largest connected region as the tumor region. The automatic segmentation model exhibited DSC of 0.68±0.14, and the semiautomatic segmentation model showed DSC of 0.61±0.23. The automatic segmentation model was superior to the semiautomatic segmentation model (P=0.035). As automated segmentation models continue to advance, they will offer substantial assistance in clinical practice.
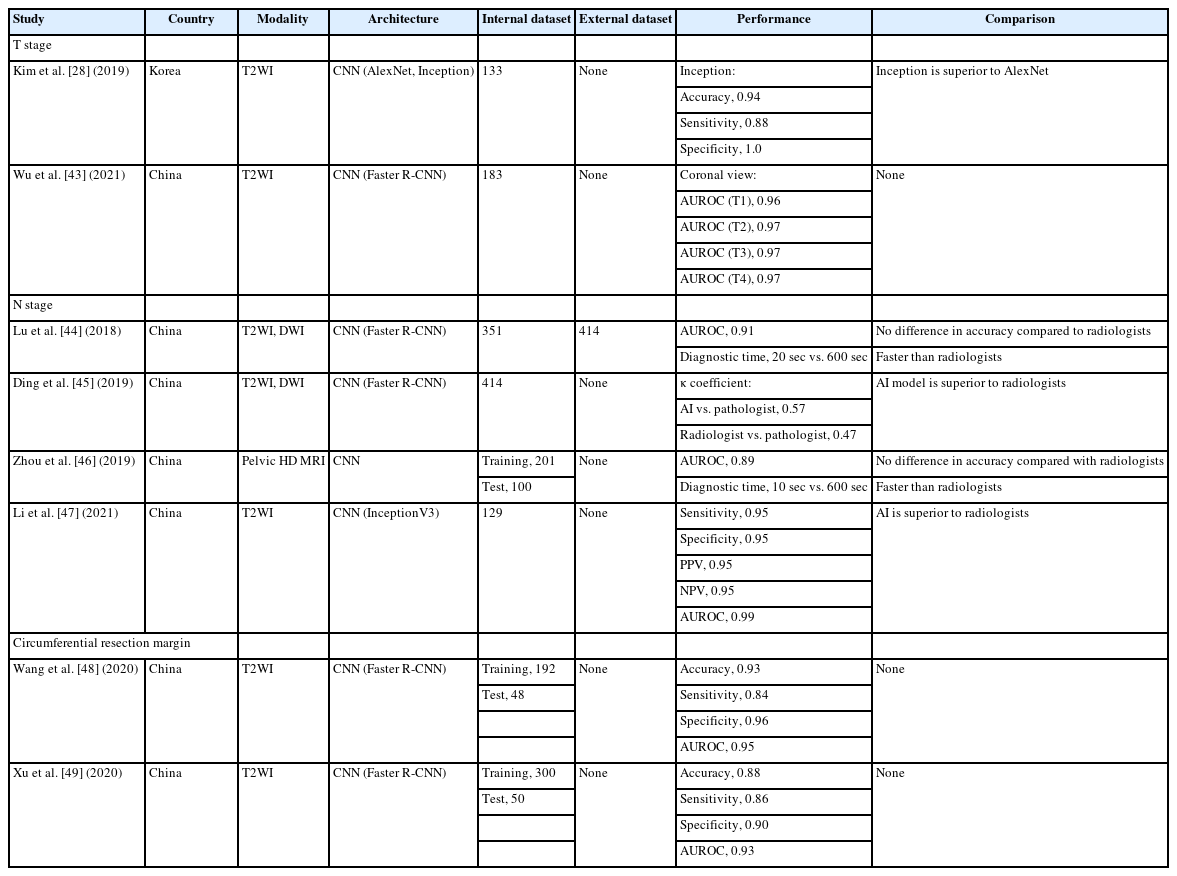
Staging
According to current guidelines for the treatment of rectal cancer, accurate staging is essential. MRI is considered the optimal modality for local tumor and lymph node staging of rectal cancer [16, 23, 28]. The predictive accuracy of pathological T categorization of rectal cancer using MRI has been reported to be approximately 71% to 91% [28, 41]. MRI evaluates lymph node status by measuring the short axial diameter and can achieve 58% to 70% sensitivity and 75% to 85% specificity for identifying metastasis [42]. However, a surplus of imaging data coupled with a shortage of radiologists has led to an increased workload for radiologists. This has led to highly stressful environments and disparities in diagnostic accuracy among radiologists. MRI interpretations are influenced by radiologists, occasionally leading to misdiagnoses [43].
Table 2 summarizes AI studies on the staging of rectal cancer [28, 43–49]. AI has been rapidly applied in MRI-based T staging, and most studies have focused on binary classification to distinguish between T1–T2 and T3–T4 to determine whether preoperative CRT should be performed. Kim et al. [28] used U-Net [36], AlexNet [50], and InceptionV3 [51] to discriminate between T2 and T3 based on the MR images obtained from 133 patients with rectal cancer. These models performed better when the input images were used as a tumor segmentation map. The inception model exhibited the best performance, with an accuracy of 0.94, sensitivity of 0.88, and specificity of 1.00. Wu et al. [43] used a Faster R-CNN model [52] for T staging based on T2WI obtained from 183 patients with rectal cancer. After 50 epochs of learning, the diagnostic performances were AUROC of 0.95 to 1.00 for the T1, T2, T3, and T4 stages in horizontal, sagittal, and coronal planes.
MRI offers limited semantic diagnostic clues for lymph node metastasis, such as the size, shape, and margins of lymph nodes, which are insufficient for the precise diagnosis of N staging in rectal cancer. The prediction of lymph node status has recently become a significant area of interest in recently [23]. Several studies have been conducted using the Faster R-CNN model [52] for N staging [44, 45]. Lu et al. [44] reported that AI-based staging of lymph node metastasis is accurate and fast. After a 4-step iteration training of the Faster R-CNN using T2WI and DWI obtained from 351 patients with rectal cancer, 414 patients with rectal cancer from other institutions were tested to verify the diagnostic performance. The AUROC was 0.91, and the diagnostic time was 20 seconds, which surpassed that of the radiologist (10 minutes). Ding et al. [45] used a Faster R-CNN [52] to detect lymph node metastases and predict prognosis. The T2WI and DWI of 414 patients with rectal cancer were analyzed using the AI model, and the results were compared with those of radiologists and pathologists. The correlation between the AI model and radiologists was 0.91, that between the AI model and pathologists was 0.45, and that between the radiologists and pathologists was only 0.13. The κ coefficient for N staging between the AI model and pathologists was 0.57, and that between the radiologists and pathologists was 0.47. The AI model was superior to the radiologists in terms of N staging, but its accuracy was inferior to that of the pathologists. Zhou et al. [46] used another CNN model to diagnose lymph node metastases based on pelvic MRI in 301 patients with rectal cancer. The accuracy of the AI model was not different from that of the radiologists; however, the AI model was much faster than the radiologists by 10 and 600 seconds, respectively. Li et al. [47] used the InceptionV3 model [51] for the recognition and detection of the lymph node status via transfer learning based on T2WI obtained from 129 patients with rectal cancer. The sensitivity, specificity, PPV, and negative predictive value (NPV) were 0.95, 0.95, 0.95, and 0.95, respectively. The AUROC was 0.99. The diagnostic performance of the AI model was compared with that of 2 radiologists, and it was found to be superior to the radiologists in all aspects. The more advanced the image diagnosis system based on AI models, the more efficient, accurate, and stable the staging system, and the errors caused by differences in radiologists’ diagnostic abilities will be reduced to a certain extent.
Several characteristics of rectal cancer, such as involvement of the CRM, may influence recurrence and metastasis [18, 23]. An accurate preoperative diagnosis of these factors may be helpful in establishing a tailored treatment plan. MRI is regarded as the best examination method for evaluating the involvement of CRM, with a specificity of 0.94; however, it requires considerable experience and time because of the large amount of imaging data [48, 53]. An AI model for the diagnosis of positive CRM may provide a reliable solution. Wang et al. [48] used a Faster R-CNN model [52] based on T2WI obtained from 240 patients with rectal cancer. The proportion of positive and negative CRM was 1:2. The accuracy, sensitivity, and specificity of the model were 0.93, 0.84, and 0.96, respectively. The AUROC was 0.93. Xu et al. [49] also used a Faster R-CNN model [52] based on T2WI obtained from 350 patients with rectal cancer. The accuracy, sensitivity, specificity, PPV, and NPV of this model were 0.88, 0.86, 0.90, 0.81, and 0.93, respectively. The AUROC was 0.93. The automatic recognition time of the model for a single image was 0.2 seconds. Thus, AI has the potential to predict the risk factors for rectal cancer and may be a good tool for personalized treatment strategies.
Genotyping
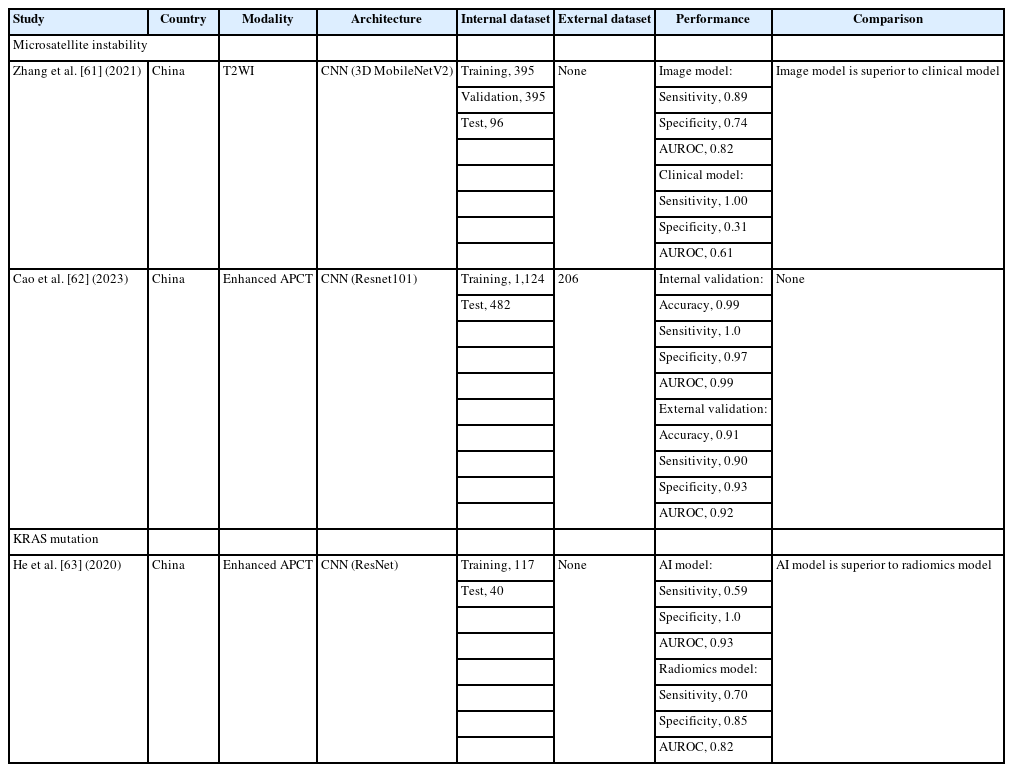
The US National Comprehensive Cancer Network (NCCN) and European Society for Medical Oncology (ESMO) guidelines recommend that all patients with rectal cancer should be tested for microsatellite instability (MSI) and KRAS mutations to establish individualized treatment strategies, thus maximizing the benefits for patients with rectal cancer [25, 54, 55]. Invasive colonoscopic biopsies or surgical specimens are essential for genetic testing using immunohistochemistry- or polymerase chain reaction (PCR)-based assays in clinical practice [56, 57]. However, this approach has several limitations. Genetic testing is not commonly performed due to its tedious procedures and heavy financial burden [58]. Moreover, the sampling procedure has potential complications [59]. Sampling errors may exist because of insufficient or tumor heterogeneity [60]. Therefore, noninvasive, feasible, low-cost, and timely methods to identify genetic mutations in rectal cancer have aroused widespread use [23].
Table 3 summarizes AI studies on genetic mutations in rectal cancer [61–63]. MSI, a consequence of the loss of one of the more mismatch repair genes, has gained considerable attention because of its significance in rectal cancer prognosis and treatment. MSI has demonstrated no benefit from 5-fluorouracil based adjuvant chemotherapy [64, 65]. More importantly, recent studies have demonstrated MSI is a predictive biomarker for immunotherapy [66, 67]. Zhang et al. [61] used the 3D MobilenetV2 model [68] based on T2WI obtained from 491 patients with rectal cancer to predict the MSI status. The performance of the AI model was compared with that of a clinical model, which is a multivariate binary logistic regression classifier based on clinical characteristics. The sensitivity and specificity of the AI model was 0.89 and 0.74, respectively. The AUROC of the AI model was 0.82. The sensitivity and specificity of the clinical model were 1.0 and 0.31, respectively. The AUROC of the clinical model was 0.61. The imaging model is superior to the clinical model in predicting the MSI status. Cao et al. [62] used the Resnet101 model [69] based on enhanced abdominopelvic computed tomography obtained from 1,606 patients with CRC to develop an AI model and from 206 patients with CRC for external validation. The AI model achieved a 0.99 accuracy, 1.0 sensitivity, and 0.97 specificity in the internal validation, and achieved 0.91 accuracy, 0.90 sensitivity, and 0.93 specificity in the external validation. For internal and external validations, the AUROC of the AI model was 0.99 in internal validation and 0.92 in external validation.
KRAS is a small G-protein that plays a role in the epidermal growth factor receptor (EGFR) pathway. Patients with rectal cancer and the KRAS mutant type show a lower response to anti-EGFR monoclonal antibodies and worse prognosis [55]. He et al. [63] used the ResNet model [69] based on enhanced abdominopelvic computed tomography images obtained from 157 patients with CRC. The diagnostic performance of the ResNet model was compared to that of a radiomics model using a random forest classifier. The ResNet model achieved a sensitivity of 0.59, specificity of 1.0, and AUROC of 0.93. The radiomics model achieved a sensitivity of 0.7, a specificity of 0.85, and an AUROC of 0.82. The ResNet model showed a superior predictive ability.
Response to therapy
Neoadjuvant CRT is a critical treatment strategy for locally advanced rectal cancer. Following CRT, 15% to 27% of patients undergo surgery despite achieving a pathological complete response (pCR) [70]. For such patients, it remains a challenge to decide whether to perform TME, which is associated with significant complications and morbidity. Various studies have shown that patients who achieve a pCR have significantly reduced local recurrence rates. Consequently, less invasive treatment options such as sphincter-saving local excision and “watch-and-wait” approaches are becoming favored in clinical practice [71, 72]. MRI plays an essential role in identifying tumor regression grade (TRG) and predicting pCR [15]. Recently, the development of radiomics and deep learning based on MRI has demonstrated impressive results in the prediction of pCR or good response (GR), defined by downstaging to ypT0–1N0 or TRG0–1 [23, 73–76]. However, these studies involved handcrafted segmentation, manual labelling, and feature definition without any deformability [77, 78]. This review focuses on image-based AI models for predicting pCR or GR. Table 4 summarizes the AI studies for predicting treatment response after neoadjuvant chemoradiotherapy in rectal cancer [78–81]. Shi et al. [79] used a CNN based on T2WI obtained from 51 patients with rectal cancer, both before and after 3 or 4 weeks of CRT. The performance of the CNN model was compared to that of the radiomics model. For predicting pCR, the AUROC of the CNN model was 0.83 and that of the radiomics model was 0.81. For predicting GR, the AUROC of the CNN model was 0.74 and that of the radiomics model was 0.92. There were no differences in predicting pCR; however, the CNN model was inferior to the radiomics model in predicting GR (P=0.04). Zhang et al. [80] used a CNN model based on T2WI obtained from 290 patients with rectal cancer, both before and after CRT. The performance of the CNN model was evaluated using T2WI obtained from 93 patients at external institutions. The accuracy, sensitivity, specificity, and AUROC of predicting pCR were 0.98, 1.0, 0.97, and 0.99, respectively. Zhu et al. [81] used a CNN model based on pre-CRT T2WI obtained from 700 patients with rectal cancer. The sensitivity, specificity, and AUROC for predicting the GR were 0.93, 0.62, and 0.81, respectively. Jang et al. [78] used ShuffleNet [82] based on the post-CRT T2WI obtained from 466 patients with rectal cancer. The prediction performance of the AI model was compared with that of senior radiologists and radiation oncologists. The accuracy, sensitivity, and specificity of the AI model for predicting a pCR were 0.85, 0.3, and 0.96, respectively. The accuracy, sensitivity, and specificity of GR prediction of the AI model were 0.72, 0.54, and 0.81, respectively. The AI model had superior predictive performance compared to radiologists and radiation oncologists. Thus, an image-based AI model for predicting treatment response has the potential to help establish tailored treatment strategies for patients with locally advanced rectal cancer.
DISCUSSION
In this systematic review, most image-based AI studies for rectal cancer research focused on tumor segmentation, staging, and treatment response. According to our review, these parameters, which could have heterogeneity according to the diagnostic tool or a radiologist’s interpretations, still require human effort over time, such as manual annotation rather than automatization. Furthermore, we hypothesized that the diagnostic performance of AI model could surpass that of humans within a brief timeframe, as indicated in some studies. In the era of a plethora of AI algorithms, there has been a recent emphasis on the importance of the performance reporting design of AI research. Simultaneously, the clinical application and approval of AI technologies tend to overhype their significance, necessitating considerations not only of ethical issues but also of stringent conditions such as the US Food and Drug Administration (FDA) 501(k) clearance [83]. In this study, we recognize that the current status of image-based AI studies for rectal cancer might be limited owing to various issues, including the sample size of the dataset, robust validation such as external validation or randomized control studies in comparison with conventional algorithms or human assessment, automated processing, and prognostic relevance based on the analysis of survival outcomes.
According to our systematic review, the sample size of the test dataset for performance was <100 in most studies. The key to improving the performance of the AI model is only obtaining more data; if possible, it is better to have a standard reference, preferably based on normal cases, for a robust dataset. However, large datasets collected from multiple institutions have some pitfalls in terms of data heterogeneity and noise, which might induce bias or loss of function with the need for a normalization process. The process of distinguishing signals from noise has become more challenging over time. A major push towards unsupervised learning techniques might enable the full utilization of vast archives and deal with difficulties in curating and labelling data. Moreover, various investigators experienced hurdles to external data exfiltration because of the particular approval for data exfiltration. This could hamper multicenter study design and even external validation with a risk for low-quality and low-quantity data. In this systematic review, we found a few multicenter studies. However, these aspects are helpful and mandatory for the clinical use of AI devices. Recently, despite the change towards non-handcrafted engineering, without human interference, a few image-based AI studies for rectal cancer showed automatization to yield performance in this field. Although the benefit of automatization over handcrafted segmentation remains unclear, an automatized process for performance might be challenging in image-based study of rectal cancer using neural networks.
The current TNM staging system with parameters, including T and N risk factors, CRM, and tumor response, has been demonstrated by confirmative evidence from many previous studies. We considered the absence of data from studies demonstrating the relationship between AI performance and prognosis through oncologic analyses involving survival outcomes, including overall, disease-free, or local recurrence. Furthermore, the opacity of the algorithms in the studies we included in our systematic review poses a challenge for their clinical implementation, given to the use of black-box algorithms. The prognostic significance of AI-based imaging parameters cannot be assigned to patients when clinicians lack comprehension. Therefore, the reporting system of AI research should extend beyond reporting diagnostic values such as accuracy, specificity, sensitivity, and ROC value. Instead, it should strive to elucidate the underlying reasons for predictions, aiming to enhance understanding and knowledge of the algorithm. Although such questioning is possible with explicitly programmed mathematical models such as conventional algorithms, neural networks based on deep learning have opaque inner workings [20]. Furthermore, in terms of image texture, feature map images displayed by AI still require more specific, clear annotation and image texture when compared to annotations for image parameters characterized by radiologists.
In conclusion, we have found that the current status of image-based AI models for rectal cancer faces multiple challenges. These include limited dataset sizes, absence of standardized references, and the difficulty for designing multicenter studies with external validation due to an insufficient dataset, despite achieving acceptable diagnostic performance. These findings suggest that the application of this model may not be feasible in clinical practice. Furthermore, an oncologic association between AI-driven classes and the prognosis of patients with rectal cancer is warranted. Overcoming pitfalls and hurdles is essential for the feasible integration of AI models into clinical settings for rectal cancer and further research based on advanced techniques, such as unsupervised learning and robust validation from high-quality labelled large datasets with standard references.
Notes
Conflict of interest
Bo Young Oh and Il Tae Son are Editorial Board members of Annals of Coloproctology, but were not involved in in the peer reviewer selection, evaluation, or decision process of this article. No other potential conflict of interest relevant to this article was reported.
Funding
None.
Author contributions
Conceptualization: ITS, BJC; Formal analysis: MK, PT; Investigation: MK, PT, MJK, BYO; Methodology: PT; Project administration: ITS, BJC; Supervision: MJK, BYO, Validation: MJK, BYO, Writing–original draft: MK; Writing–review & editing: all authors. All authors read and approved the final manuscript.